Role of genetic heterogeneity and epistasis in bladder cancer susceptibility and outcome: a learning classifier system approach
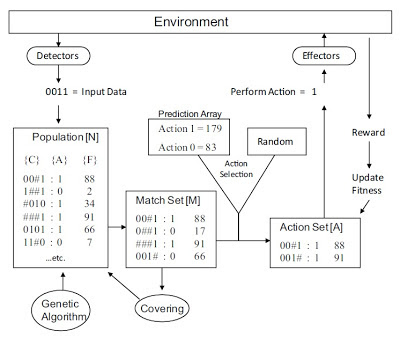
This open-access paper reports our successful application of learning classifier systems (LCS) to the genetic analysis of bladder cancer. This method is promising because it can detect both epistasis and genetic or locus heterogeneity. First read our comprehensive review of LCS. We also have a more recent overview of our own LCS approach in Computational Intelligence Magazine.
Urbanowicz RJ, Andrew AS, Karagas MR, Moore JH. Role of genetic heterogeneity and epistasis in bladder cancer susceptibility and outcome: a learning classifier system approach. J Am Med Inform Assoc 20, 603-612 (2013). [PubMed] [PDF]
Abstract
BACKGROUND AND OBJECTIVE: Detecting complex patterns of association between genetic or environmental risk factors and disease risk has become an important target for epidemiological research. In particular, strategies that provide multifactor interactions or heterogeneous patterns of association can offer new insights into association studies for which traditional analytic tools have had limited success.
MATERIALS AND METHODS: To concurrently examine these phenomena, previous work has successfully considered the application of learning classifier systems (LCSs), a flexible class of evolutionary algorithms that distributes learned associations over a population of rules. Subsequent work dealt with the inherent problems of knowledge discovery and interpretation within these algorithms, allowing for the characterization of heterogeneous patterns of association. Whereas these previous advancements were evaluated using complex simulation studies, this study applied these collective works to a 'real-world' genetic epidemiology study of bladder cancer susceptibility.
RESULTS AND DISCUSSION: We replicated the identification of previously characterized factors that modify bladder cancer risk-namely, single nucleotide polymorphisms from a DNA repair gene, and smoking. Furthermore, we identified potentially heterogeneous groups of subjects characterized by distinct patterns of association. Cox proportional hazard models comparing clinical outcome variables between the cases of the two largest groups yielded a significant, meaningful difference in survival time in years (survivorship). A marginally significant difference in recurrence time was also noted. These results support the hypothesis that an LCS approach can offer greater insight into complex patterns of association.
CONCLUSIONS: This methodology appears to be well suited to the dissection of disease heterogeneity, a key component in the advancement of personalized medicine.


0 Comments:
Post a Comment
<< Home